Research
The Chair of Biochemistry IV aims to unravel the hidden secrets behind the central dogma of molecular biology. While it is commonly known that there is much more beyond the DNA-to-RNA-to-protein rule, the manifold mechanisms behind the regulation of gene expression are far from being understood. We have contributed and we will continue to contribute essential information on how general RNA-binding proteins and long non-coding RNAs regulate transcription, 3’ end mRNA processing and translation.
A key model system, which allows us to understand these intricate processes is Drosophila dosage compensation (see here for more information). The information our research group has obtained during the last decade allows us now to venture into the human system, where we establish a link between cellular function and various diseases.
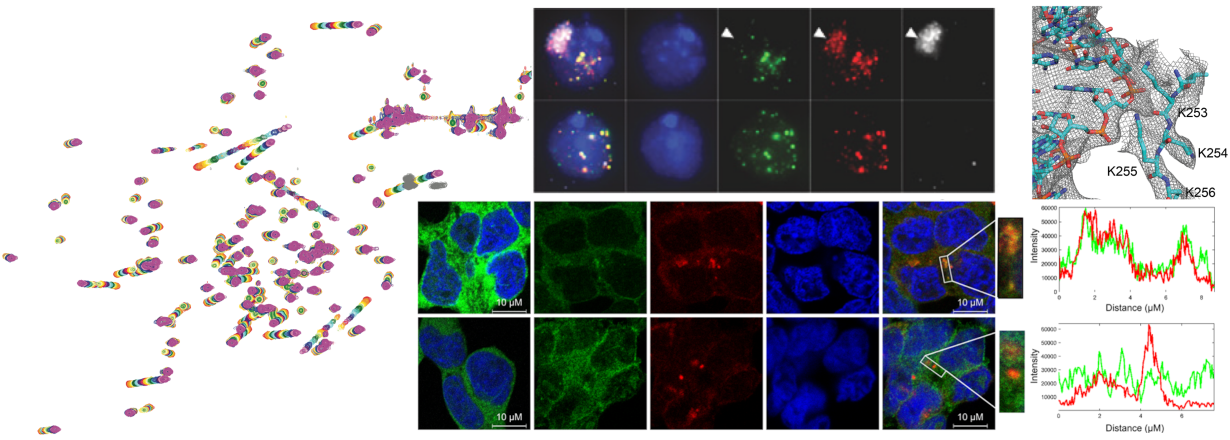
Another focus of the lab is to understand the link between the ubiquitin system and RNA binding. The modification of proteins with the small protein ubiquitin by covalent linkage has profound effects on all aspects of the human cell. E3 ligase enzymes actively support this ubiquitin modification and convey substrate selectivity. A certain subfamily of E3 ligases, the tripartite motif (TRIM) protein family has been shown to be highly important in innate immunity and antiviral defence mechanisms of the cell. Some of these have been shown to bind RNA and we want to understand what influence RNA binding has on the protein function and cellular fate. We could show that RNA binding of TRIM25 is more important for its antiviral defence than its E3 ligase activity after infection with a range of viruses.
To answer our biological questions, we need to adapt our methods portfolio continuously and are therefore involved in developing integrative structure modelling approaches, tailor-made for each system we study.
With well-equipped molecular and biochemistry labs and access to a full range of high-flield solid- and liquid-state NMR spectrometers (600 MHz, 700 MHz, 900 MHz and 1 GHz) located at the NBNC (Northern Bavarian NMR Center), the Chair of Biochemistry IV is able to study structure and dynamics of biomolecular complexes. Frequent access to synchrotron beam lines and cryo-EM microscopes enables our approach of integrative structure modelling of biomolecular complexes (see Methods). Thus, we are enabled to venture into the dark matter of structural biology: intrinsically disordered proteins, RNA structure and 4D structural biology (time and space).
For details see:
→ Research AG Hennig
→ Research AG Jagtap
→ Research AG Soni